Daniel P Neumann
Molecular Biologist
Greetings

About me
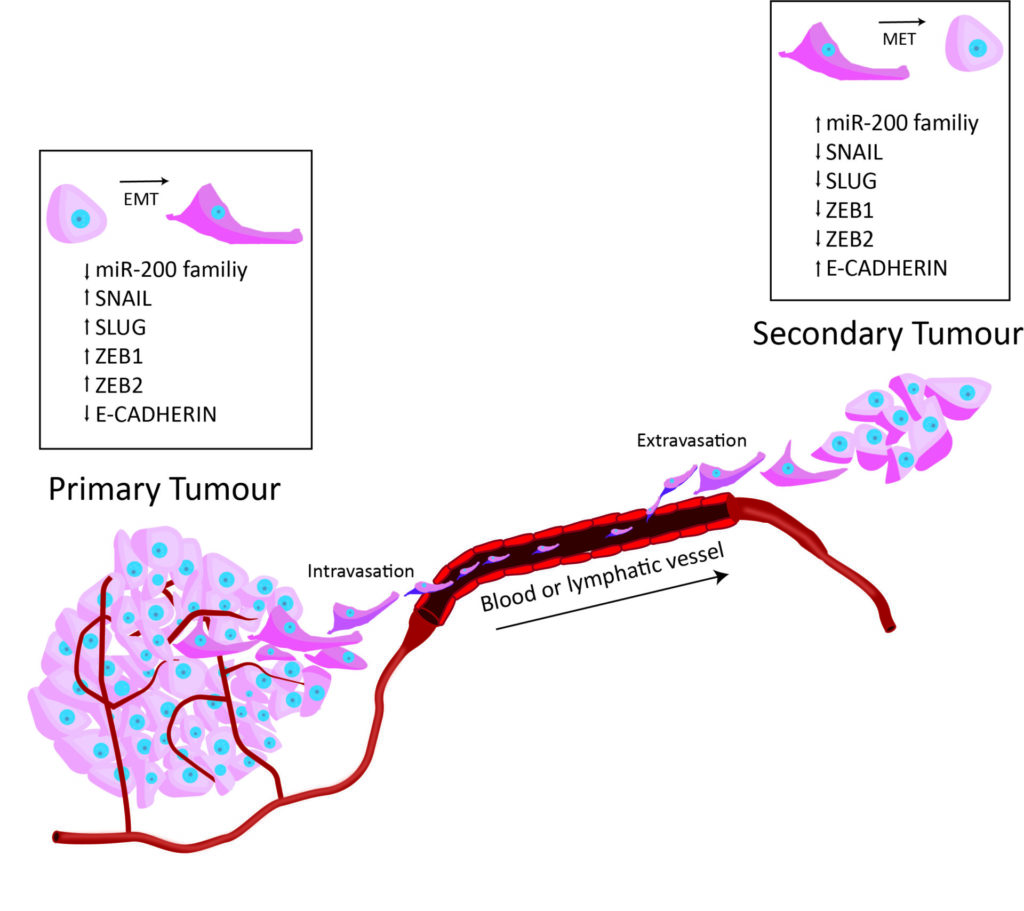
- I am a molecular biologist from South Australia who recently defended my PhD, The role of RNA-binding proteins in epithelial-mesenchymal transition.
- My broad interests include gene regulation, synthetic biology and CRISPR, single-cell and spatial transcriptomics, cell differentiation and fate determination, breast cancer and prostate cancer.
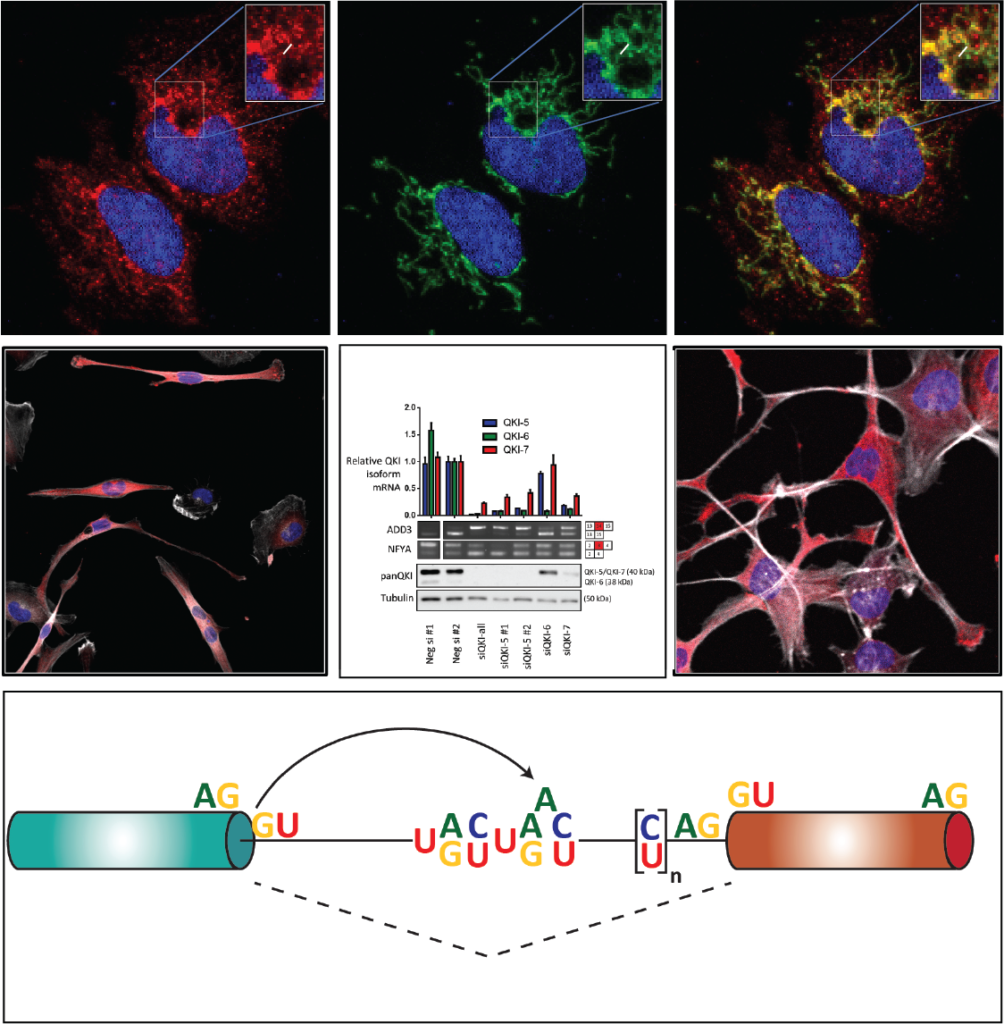
- I am currently studying how the RNA-binding protein, QKI, regulates the alternative splicing of its own transcript to produce its isoforms, which intend on publishing shortly.
- I work as a wet lab scientist and bioinformatician.
- My passions outside of work include composing music and playing piano and guitar.
Wet Lab Techniques
- Endpoint Polymerase Chain Reaction (PCR) and REAL-TIME quantitative-PCR
- Western blot and SDS-PAGE
- Transwell migration and invasion assays
- Experienced in both phase and confocal microscopy
- Combination of fluorescence microscopy and imagej macros to assess changes in cell morphology and nuclear/cytoplasmic localization of proteins
- Incucyte proliferation and scratch wound assays
- microRNA and siRNA cell transfections
- Molecular cloning and generation of expression constructs
- Mammalian Cell culture techniques
- Generating viral constructs and transducing cells
- CRISPR Cas9 mediated gene knockout in human cells
Bioinformatics Techniques
- Confident programming in Python with some experience in R.
- Proficient usage of the Linux command line, bash scripting and usage of bioinformatic software.
- Implementation of various methods of survival, statistical and gene expression analyses on public datasets including the cancer genome atlas (TCGA), cancer cell line encyclopaedia (CCLE), gene-tissue expression (GTEx) portal, and the Chinese glioma genome atlas (CGGA).
- Experience in many Python modules including pandas, SciPy, NumPy Matplotlib, Seaborn, Bokeh and pyGenomeTracks.
- Some experience in processing NGS and RNA-Seq data, particularly in the context of HITS-CLIP analysis.
- Motif analysis using MEME, HOMER or regular expressions.
Publications
Neumann, D.P., Goodall, G.J. and Gregory, P.A., 2018, March. Regulation of splicing and circularisation of RNA in epithelial mesenchymal plasticity. In Seminars in cell & developmental biology (Vol. 75, pp. 50-60). Academic Press.
Pillman, K.A., Phillips, C.A., Roslan, S., Toubia, J., Dredge, B.K., Bert, A.G., Lumb, R., Neumann, D.P., Li, X., Conn, S.J., Liu, D., Bracken, C.P., Lawrence, D.M., Stylianou, N., Schreiber, A.W., Tilley, W.D., Hollier, B.G., Khew-Goodall, Y., Selth, L.A., Goodall, G.J., & Gregory, P.A. (2018). miR-200/375 control epithelial plasticity-associated alternative splicing by repressing the RNA-binding protein Quaking. The EMBO journal, e99016
Conferences
- Presented at the 3rd Indo-Australia Symposium on Epithelial-Mesenchymal Transition (November 2019)
- Presented at the Centre for Cancer Biology Inaugural Student and
Early-Mid Career Researcher Symposium (July 2019) - Presented a poster for the Adelaide Protein Group Student Awards (June 2019)
- Presented a poster for the 2nd Japan-Australia joint RNA (JAJ RNA) meeting at Hokkaido University,
Sapporo, Japan (November 2018) - Presented a poster at the ComBio meeting at the Adelaide Convention Centre (October 2017)
- Presented at the South Australian Men’s Health Research Symposium at SAHMRI (June 2015)
- Presented at the 2015 ASMR South Australian scientific meeting (May 2015)